Research Tracks
Can we engineer microbial communities to promote food quality and security, and to optimize functional roles in human digestion and health? The Food Systems Biotechnology group develops computational methods and utilizes "omics" technologies to study microbial ecosystems in food production and human health, with the ultimate goal of creating microbial communities and products that optimize food quality, safety, and human health.
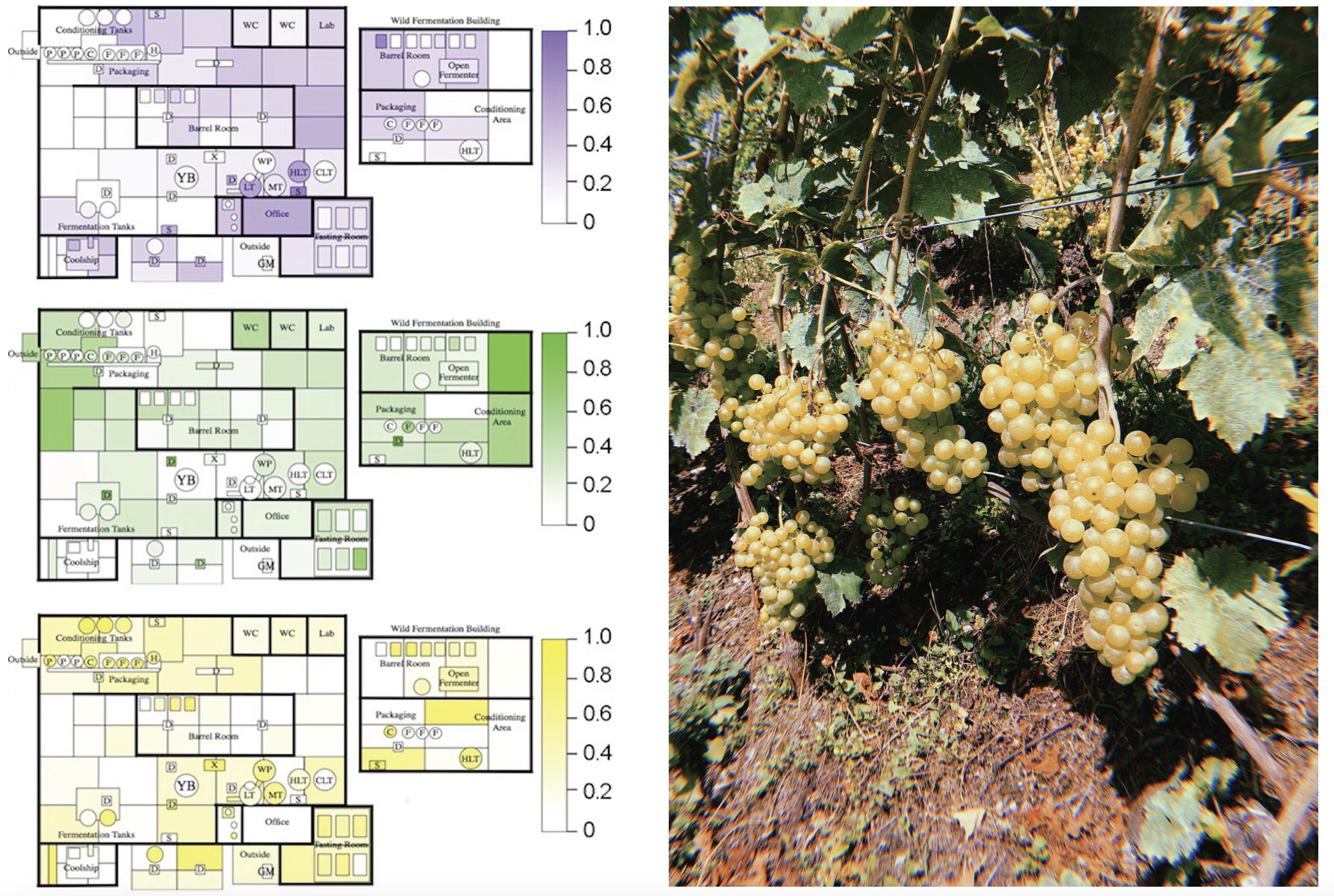
Microbial communities impact food quality and safety at all stages of the food production chain. The Food Systems Biotechnology group utilizes integrated "omics" approaches to examine the interconnection of biotic and abiotic factors, microbiomes, and characteristics of regional food products and food production systems, with the ultimate goal of reverse engineering these systems to improve food quality, safety, and sustainability. This includes characterizing microbial properties of food products that underpin regional quality characteristics ("microbial terroir"), as well as studying the temporospatial organization of microbiomes in food production systems both in the field and during food processing, to better predict and control microbial activity during food production.
Microbial communities are shaped by host factors, environmental conditions, and human influences, and have a strong impact on food quality and safety at every stage of the food production chain. This is particularly of interest in vineyards, since regionally varying factors, including associated microbiota (microbial terroir), produce wines with distinct sensory qualities. Yet the complex interactions underpinning these local differences are poorly understood. Therefore, the Food Systems Biotechnology Group utilizes integrated "omics" approaches to disentangle biotic and abiotic factors and to gain a more holistic insight into plant-microbiome-environment interactions. This includes characterizing microbial properties of food products that underpin regional quality characteristics ("microbial terroir"), as well as studying the temporospatial organization of microbiomes in food production systems both in the field and during food processing, to better predict and control microbial activity during food production.
This includes investigation of microbial ecosystems of wine production, encompassing both vineyard and winery environments, and the role of climate, human management practices, and plant genotype in regulating the microbial and chemical composition of wine and other foods; factors that can be harnessed for quality improvement. This same model is applied to other fermented foods on a broad scale, using both culture-independent untargeted “omics” technologies, as well as high-throughput culturomics for microbial isolation and characterization.
The human body hosts trillions of microbial cells (outnumbering human cells!), forming complex microbiomes that are intimately tied to human health and disease across different body sites. The Food Systems Biotechnology group utilizes integrated multi-omics approaches to study the assembly, development, and dynamics of these microbial communities, aiming to achieve a predictive understanding of their behavior and interactions with the human body. Our research focuses on the establishment of gut microbiota during infancy and childhood, the interaction of the gut microbiome with diet, the role of the microbiome in microenvironment and carcinogenesis at different human body sites, and the impacts of lifestyle and associated exposures on the gut microbiome and human health.
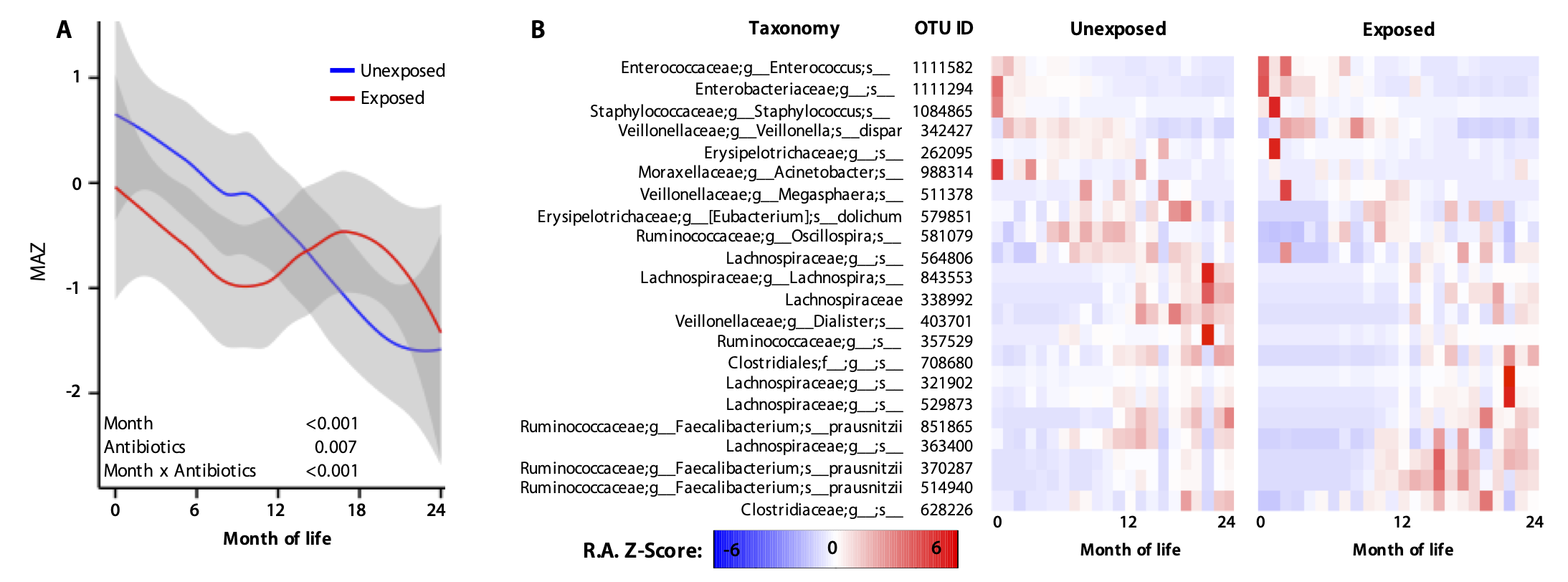
Microbiome development during early childhood
The first years of life are marked by rapid microbiome development, transitioning from being essentially sterile at birth to reaching a stable, adult-like composition. This maturation of the microbiome parallels critical milestones in physiological, immunological, and neurobehavioral development, with growing evidence of bidirectional interactions between microbiome development and host processes. We develop and utilize multi-omics and machine learning methods to investigate microbiome and host development in early life. We study how early life factors, such as antibiotic exposure, shape the developmental trajectory of the infant gut microbiome. Our research particularly focuses on the longitudinal causal relationships between functional microbiome changes and host developmental, behavioral, and health outcomes.This includes neurodevelopmental and immunological outcomes. Ultimately, we aim to establish a predictive understanding of how early life microbiomes shape childhood health, and the role that biotherapeutics could play in predicting and treating childhood diseases.
Microbiome and cancer treatment
Numerous studies have implicated the human microbiome in both cancer risk and treatment. We investigate microbiome changes in different solid and liquid cancers to identify novel biomarkers and therapeutic targets. In collaboration with clinical researchers, we employ empirical and computational approaches to study interactions between the gut/skin microbiomes, metabolomes, and personalized responses to allogeneic hematopoietic stem cell transplantation (allo-HSCT). Allo-HSCT is the only effective treatment for certain blood cancers and disorders. However, while life-saving, the procedure carries a high risk of complications, including infections and graft-versus-host-disease. Our research aims to provide a predictive understanding of how the human gut microbiome interacts with the immune system and influences morbidity risks in allo-HSCT. By identifying individualized features affecting patient response, we aim to optimize drug treatments and improve patient survival rate following allo-HSCT.
Microbiome responses to lifestyle and environmental change
Ecosystems worldwide are facing unprecedented biodiversity loss due to climate change, land use change, environmental pollution, and other human-driven impacts linked to industrialization. This includes a decline in microbial biodiversity, which is crucial for both human and planetary health. The gut microbiome, which co-evolved with humans over thousands of years to play a key role in human physiology and health, is also vulnerable to the effects of industrialization. Through diverse international collaborations, we investigate the complex responses of the human microbiome to environmental change. Our research covers a range of topics, including diet-microbiome interactions across diverse human populations, connections between the ‘exposome’ and human microbiome, and other changes in host-microbiome interactions associated with an industrialized lifestyle and its exposures. We employ a combination of sequencing and metabolomics technologies, microbiome cultivation, diverse computational approaches, and global meta-analyses to map the ecological impacts of ongoing environmental changes on microbial diversity and their functional implications for human health. Our goal is to contribute to cataloging and preserving the genetic and biosynthetic diversity of global microbiomes.
Advances in DNA sequencing technologies enable acquisition of metagenomic datasets at an unprecedented scale. More accurate methods are needed to analyze and interpret these high-dimensional data, and integrate "omics" datasets to gain detailed insight into host–microbial interactions. The Food Systems Biotechnology group develops open-source bioinformatics software and machine-learning methods to explore microbiomes and other omics datasets in diverse ecosystems, including development of diverse plugins for the QIIME 2 (external page https://qiime2.org) microbiome multi-omics platform. This includes open-source bioinformatics tools for genome and metagenome assembly, enabling in silico reconstruction of whole microbial genomes to provide information about species composition and functional genetic potential of complex microbiomes. Current activities include development of open-source software packages for infectious disease and microbial biosurveillance using shotgun metagenome sequence data, traceable and reproducible bioinformatics workflows, machine learning approaches for microbial classification and longitudinal prediction, and software for reproducible FAIR reuse of open research datasets. To see our latest projects visit external page https://github.com/bokulich-lab.
